Publications
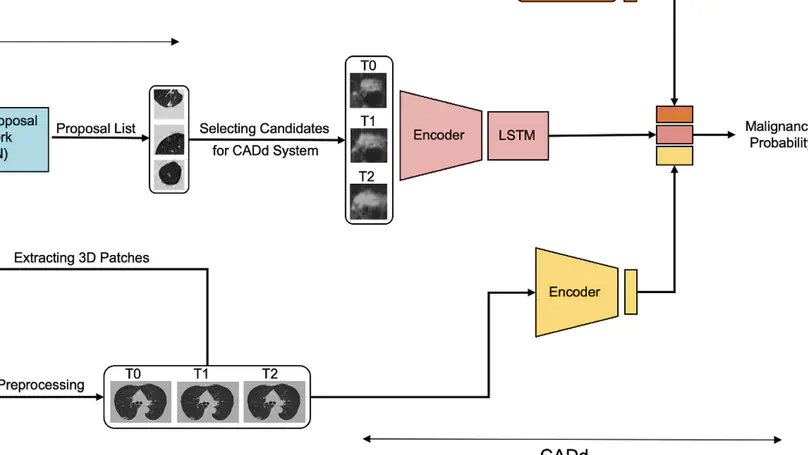
Lung cancer screening (LCS) using annual computed tomography (CT) scanning significantly reduces mortality by detecting cancerous lung nodules at an earlier stage. Deep learning algorithms can improve nodule malignancy risk stratification. However, they have typically been used to analyse single time point CT data when detecting malignant nodules on either baseline or incident CT LCS rounds. Deep learning algorithms have the greatest value in two aspects. These approaches have great potential in assessing nodule change across time-series CT scans where subtle changes may be challenging to identify using the human eye alone. Moreover, they could be targeted to detect nodules developing on incident screening rounds, where cancers are generally smaller and more challenging to detect confidently. Here, we show the performance of our Deep learning-based Computer-Aided Diagnosis model integrating Nodule and Lung imaging data with clinical Metadata Longitudinally (DeepCAD-NLM-L) for malignancy prediction. DeepCAD-NLM-L showed improved performance (AUC = 88%) against models utilizing single time-point data alone. DeepCAD-NLM-L also demonstrated comparable and complementary performance to radiologists when interpreting the most challenging nodules typically found in LCS programs. It also demonstrated similar performance to radiologists when assessed on out-of-distribution imaging dataset. The results emphasize the advantages of using time-series and multimodal analyses when interpreting malignancy risk in LCS.
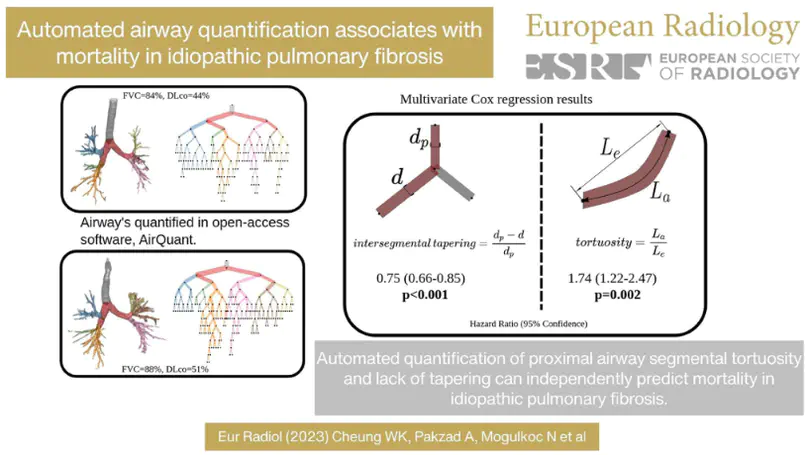
Patients with lung damage by scarring typically have abnormally larger airways. This difference in airway size between patients and healthy individuals can be seen with 3D xrays, known as a CT scan. We present a new software program called AirQuant to measure all airways of a patient’s CT scan. We compare measurements between 14 healthy individuals and 14 diseased patients. We found that airway disease was worse in the lower lungs and furthermore their airways were also more twisted. Measurements used by AirQuant, has potential as new markers in imaging to help us understand how bad a patient’s disease is.
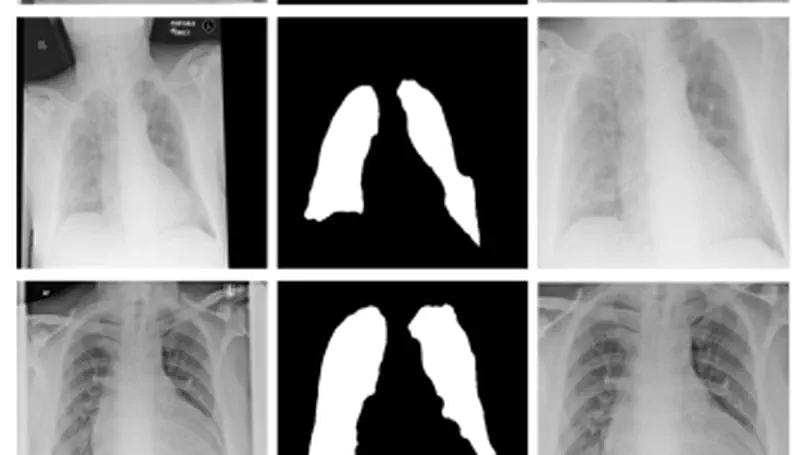
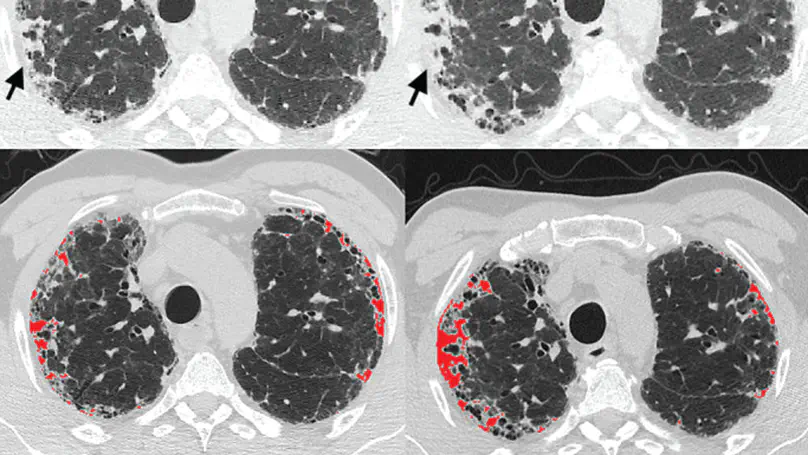
During the COVID-19 pandemic, the sheer volume of imaging performed in an emergency setting for COVID-19 diagnosis has resulted in a wide variability of clinical CXR acquisitions. This variation is seen in the CXR projections used, image annotations added and in the inspiratory effort and degree of rotation of clinical images. The image analysis community has attempted to ease the burden on overstretched radiology departments during the pandemic by developing automated COVID-19 diagnostic algorithms, the input for which has been CXR imaging. Large publicly available CXR datasets have been leveraged to improve deep learning algorithms for COVID-19 diagnosis. Yet the variable quality of clinically-acquired CXRs within publicly available datasets could have a profound effect on algorithm performance. COVID-19 diagnosis may be inferred by an algorithm from non-anatomical features on an image such as image labels. These imaging shortcuts may be dataset-specific and limit the generalisability of AI systems. Understanding and correcting key potential biases in CXR images is therefore an essential first step prior to CXR image analysis. In this study, we propose a simple and effective step-wise approach to pre-processing a COVID-19 chest X-ray dataset to remove undesired biases. We perform ablation studies to show the impact of each individual step. The results suggest that using our proposed pipeline could increase accuracy of the baseline COVID-19 detection algorithm by up to 13%.
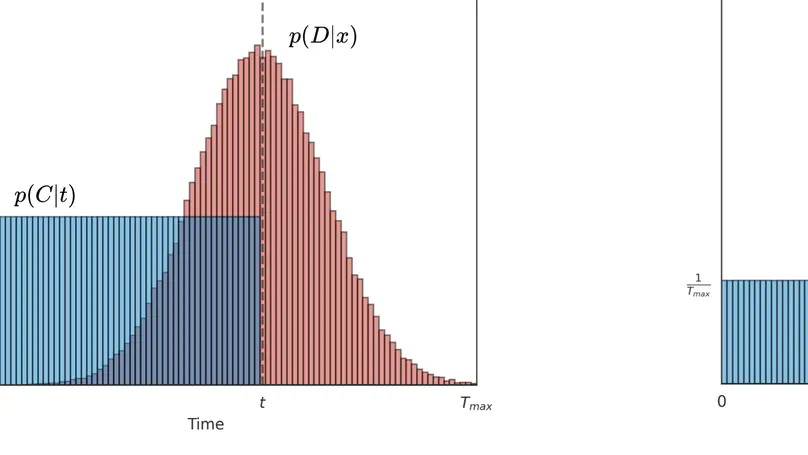
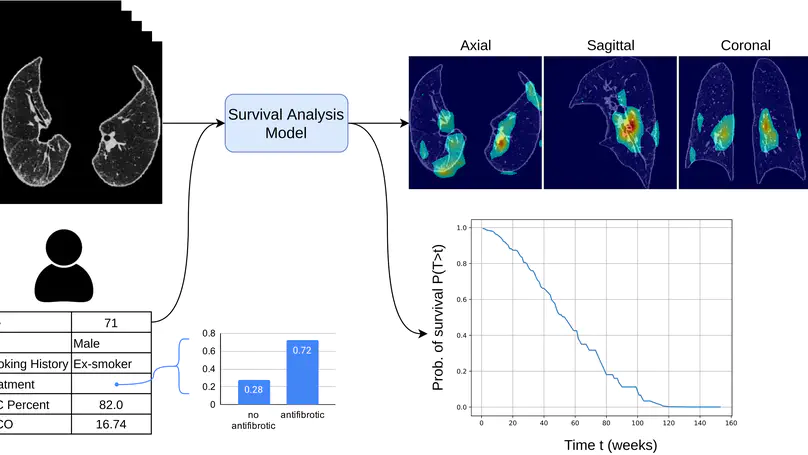
Survival analysis is a valuable tool for estimating the time until specific events, such as death or cancer recurrence, based on baseline observations. This is particularly useful in healthcare to prognostically predict clinically important events based on patient data. However, existing approaches often have limitations; some focus only on ranking patients by survivability, neglecting to estimate the actual event time, while others treat the problem as a classification task, ignoring the inherent time-ordered structure of the events. Additionally, the effective utilisation of censored samples data points where the event time is unknown is essential for enhancing the model’s predictive accuracy. In this paper, we introduce CenTime, a novel approach to survival analysis that directly estimates the time to event. Our method features an innovative event-conditional censoring mechanism that performs robustly even when uncensored data is scarce. We demonstrate that our approach forms a consistent estimator for the event model parameters, even in the absence of uncensored data. Furthermore, CenTime is easily integrated with deep learning models with no restrictions on batch size or the number of uncensored samples. We compare our approach to standard survival analysis methods, including the Cox proportional-hazard model and DeepHit. Our results indicate that CenTime offers state-of-the-art performance in predicting time-to-death while maintaining comparable ranking performance. Our implementation is publicly available at https://github.com/ahmedhshahin/CenTime.
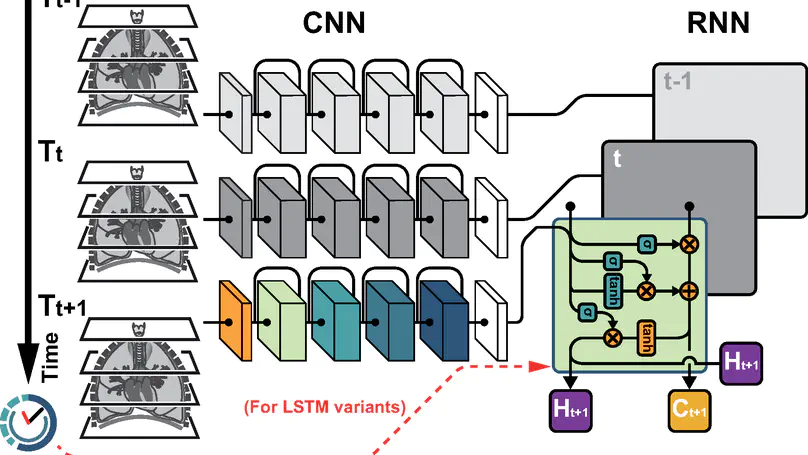
In this study, we present a hybrid CNN-RNN approach to investigate long-term survival of subjects in a lung cancer screening study. Subjects who died of cardiovascular and respiratory causes were identified whereby the CNN model was used to capture imaging features in the CT scans and the RNN model was used to investigate time series and thus global information. To account for heterogeneity in patients’ follow-up times, two different variants of LSTM models were evaluated, each incorporating different strategies to address irregularities in follow-up time. The models were trained on subjects who underwent cardiovascular and respiratory deaths and a control cohort matched to participant age, gender, and smoking history. The combined model can achieve an AUC of 0.76 which outperforms humans at cardiovascular mortality prediction. The corresponding F1 and Matthews Correlation Coefficient are 0.63 and 0.42 respectively. The generalisability of the model is further validated on an ‘external’ cohort. The same models were applied to survival analysis with the Cox Proportional Hazard model. It was demonstrated that incorporating the follow-up history can lead to improvement in survival prediction. The Cox neural network can achieve an IPCW C-index of 0.75 on the internal dataset and 0.69 on an external dataset. Delineating subjects at increased risk of cardiorespiratory mortality can alert clinicians to request further more detailed functional or imaging studies to improve the assessment of cardiorespiratory disease burden. Such strategies may uncover unsuspected and under-recognised pathologies thereby potentially reducing patient morbidity.
The COVID-19 pandemic that began in 2019 has resulted in millions of deaths worldwide. Over this period, the economic and healthcare consequences of COVID-19 infection in survivors of acute COVID-19 infection have become apparent. During the course of the pandemic, computer analysis of medical images and data have been widely used by the medical research community. In particular, deep-learning methods, which are artificial intelligence (AI)-based approaches, have been frequently employed. This paper provides a review of deep-learning-based AI techniques for COVID-19 diagnosis using chest radiography and computed tomography. Thirty papers published from February 2020 to March 2022 that used two-dimensional (2D)/three-dimensional (3D) deep convolutional neural networks combined with transfer learning for COVID-19 detection were reviewed. The review describes how deep-learning methods detect COVID-19, and several limitations of the proposed methods are highlighted.

Several chronic lung diseases, like idiopathic pulmonary fibrosis (IPF) are characterised by abnormal dilatation of the airways. Quantification of airway features on computed tomography (CT) can help characterise disease severity and progression. Physics based airway measurement algorithms that have been developed have met with limited success, in part due to the sheer diversity of airway morphology seen in clinical practice. Supervised learning methods are not feasible due to the high cost of obtaining precise airway annotations. We propose synthesising airways by style transfer using perceptual losses to train our model: Airway Transfer Network (ATN). We compare our ATN model with a state-of-the-art GAN-based network (simGAN) using a) qualitative assessment; b) assessment of the ability of ATN and simGAN based CT airway metrics to predict mortality in a population of 113 patients with IPF. ATN was shown to be quicker and easier to train than simGAN. ATN-based airway measurements showed consistently stronger associations with mortality than simGAN-derived airway metrics on IPF CTs. Airway synthesis by a transformation network that refines synthetic data using perceptual losses is a realistic alternative to GAN-based methods for clinical CT analyses of idiopathic pulmonary fibrosis. Our source code can be found at https://github.com/ashkanpakzad/ATN that is compatible with the existing open-source airway analysis framework, AirQuant.
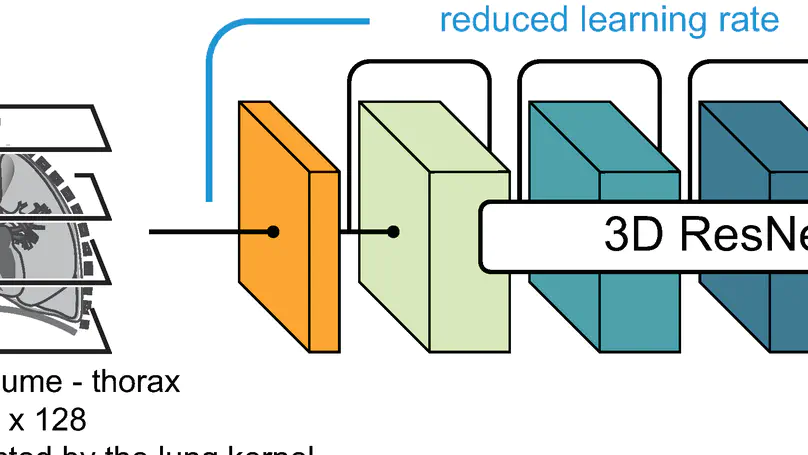
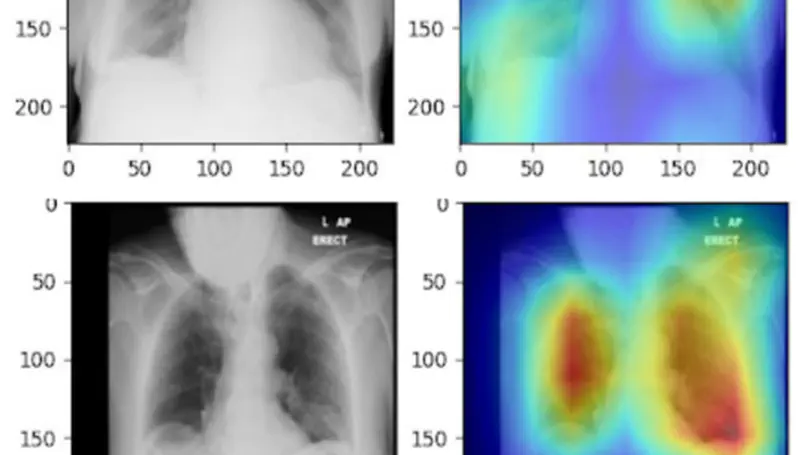
In this study, the long-term mortality in the National Lung Screening Trial (NLST) was investigated using a deep learning-based method. Binary classification of the non-lung-cancer mortality (i.e. cardiovascular and respiratory mortality) was performed using neural network models centered around a 3D-ResNet. The models were trained on a participant age, gender, and smoking history matched cohort. Utilising both the 3D CT scan and clinical information, the models can achieve an AUC of 0.73 which outperforms humans at cardiovascular mortality prediction. By interpreting the trained models with 3D saliency maps, we examined the features on the CT scans that correspond to the mortality signal. The saliency maps can potentially assist the clinicians’ and radiologists’ to identify regions of concern on the image that may indicate the need to adopt preventative healthcare management strategies to prolong the patients’ life expectancy.
Idiopathic Pulmonary Fibrosis (IPF) is an inexorably progressive fibrotic lung disease with a variable and unpredictable rate of progression. CT scans of the lungs inform clinical assessment of IPF patients and contain pertinent information related to disease progression. In this work, we propose a multi-modal method that uses neural networks and memory banks to predict the survival of IPF patients using clinical and imaging data. The majority of clinical IPF patient records have missing data (e.g. missing lung function tests). To this end, we propose a probabilistic model that captures the dependencies between the observed clinical variables and imputes missing ones. This principled approach to missing data imputation can be naturally combined with a deep survival analysis model. We show that the proposed framework yields significantly better survival analysis results than baselines in terms of concordance index and integrated Brier score. Our work also provides insights into novel image-based biomarkers that are linked to mortality.
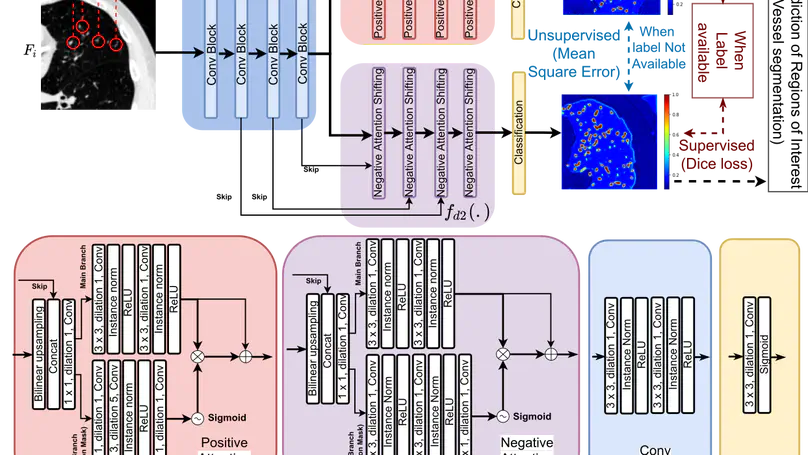
We propose MisMatch, a novel consistency-driven semi-supervised segmentation framework which produces predictions that are invariant to learnt feature perturbations. MisMatch consists of an encoder and a two-head decoders. One decoder learns positive attention to the foreground regions of interest (RoI) on unlabelled images thereby generating dilated features. The other decoder learns negative attention to the foreground on the same unlabelled images thereby generating eroded features. We then apply a consistency regularisation on the paired predictions. MisMatch outperforms state-of-the-art semi-supervised methods on a CT-based pulmonary vessel segmentation task and a MRI-based brain tumour segmentation task. In addition, we show that the effectiveness of MisMatch comes from better model calibration than its supervised learning counterpart.
Patients with lung damage by scarring typically have abnormally larger airways. This difference in airway size between patients and healthy individuals can be seen with 3D xrays, known as a CT scan. We present a new software program called AirQuant to measure all airways of a patient’s CT scan. We compare measurements between 14 healthy individuals and 14 diseased patients. We found that airway disease was worse in the lower lungs and furthermore their airways were also more twisted. Measurements used by AirQuant, has potential as new markers in imaging to help us understand how bad a patient’s disease is.
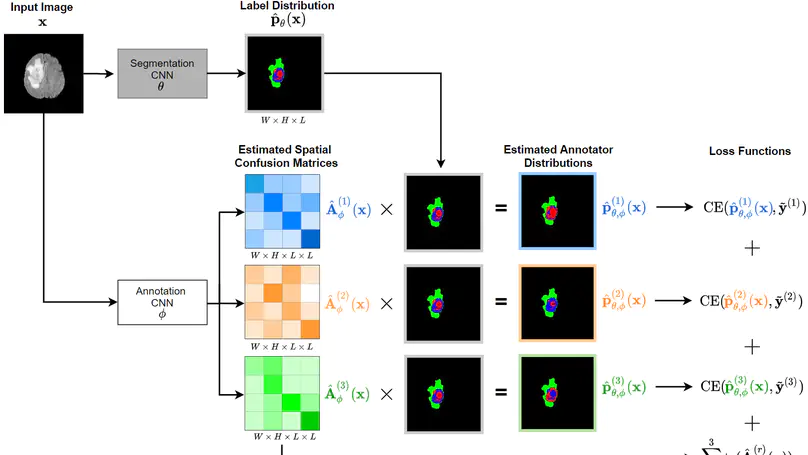
Recent years have seen increasing use of supervised learning methods for segmentation tasks. However, the predictive performance of these algorithms depends on the quality of labels. This problem is particularly pertinent in the medical image domain, where both the annotation cost and inter-observer variability are high. In a typical label acquisition process, different human experts provide their estimates of the “true” segmentation labels under the influence of their own biases and competence levels. Treating these noisy labels blindly as the ground truth limits the performance that automatic segmentation algorithms can achieve. In this work, we present a method for jointly learning, from purely noisy observations alone, the reliability of individual annotators and the true segmentation label distributions, using two coupled CNNs. The separation of the two is achieved by encouraging the estimated annotators to be maximally unreliable while achieving high fidelity with the noisy training data. We first define a toy segmentation dataset based on MNIST and study the properties of the proposed algorithm. We then demonstrate the utility of the method on three public medical imaging segmentation datasets with simulated (when necessary) and real diverse annotations: 1) MSLSC (multiple-sclerosis lesions); 2) BraTS (brain tumours); 3) LIDC-IDRI (lung abnormalities). In all cases, our method outperforms competing methods and relevant baselines particularly in cases where the number of annotations is small and the amount of disagreement is large. The experiments also show strong ability to capture the complex spatial characteristics of annotators’ mistakes. Our code is available at https: //github.com/moucheng2017/Learn_Noisy_Labels_Medical_Images.
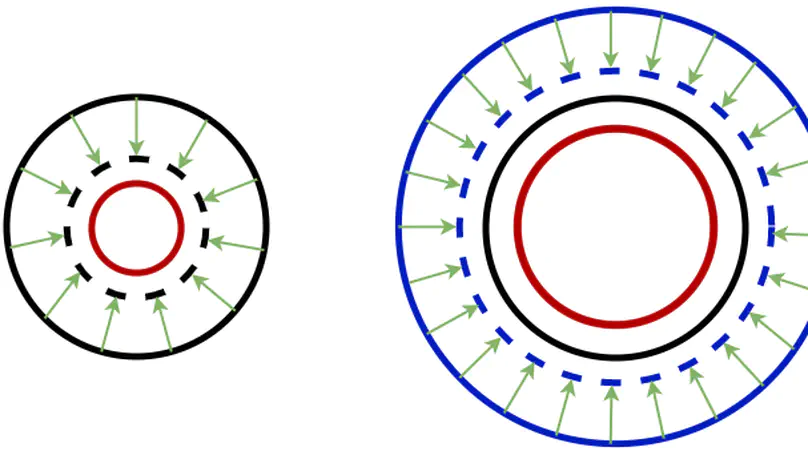
In convolutional neural network based medical image segmentation, the periphery of foreground regions representing malignant tissues may be disproportionately assigned as belonging to the background class of healthy tissues [18][21][24][12][4]. Misclassification of foreground pixels as the background class can lead to high false negative detection rates. In this paper, we propose a novel attention mechanism to directly address such high false negative rates, called Paying Attention to Mistakes. Our attention mechanism steers the models towards false positive identification, which counters the existing bias towards false negatives. The proposed mechanism has two complementary implementations: (a) “explicit” steering of the model to attend to a larger Effective Receptive Field on the foreground areas; (b) “implicit” steering towards false positives, by attending to a smaller Effective Receptive Field on the background areas. We validated our methods on three tasks: 1) binary dense prediction between vehicles and the background using CityScapes; 2) Enhanced Tumour Core segmentation with multi-modal MRI scans in BRATS2018; 3) segmenting stroke lesions using ultrasound images in ISLES2018. We compared our methods with state-of-the-art attention mechanisms in medical imaging, including self-attention, spatial-attention and spatial-channel mixed attention. Across all of the three different tasks, our models consistently outperform the baseline models in Intersection over Union (IoU) and/or Hausdorff Distance (HD). For instance, in the second task, the “explicit” implementation of our mechanism reduces the HD of the best baseline by more than 26%, whilst improving the IoU by more than 3%. We believe our proposed attention mechanism can benefit a wide range of medical and computer vision tasks, which suffer from over-detection of background.